Smart Cities
We develop, monitor, and improve infrastructure, buildings, transportation routes, the power supply, and everyday activities in crowded, urban environments.
Learn More
Our centers are engines of translational research and education in the data sciences, and a source of technology with high commercialization potential.
We develop, monitor, and improve infrastructure, buildings, transportation routes, the power supply, and everyday activities in crowded, urban environments.
Learn More
We study the physical aspects of sensing, generating, collecting, storing, transporting, and processing large data sets.
Learn More
We work to improve the health of individuals and the health care system through data-driven methods and understanding of health processes.
Learn More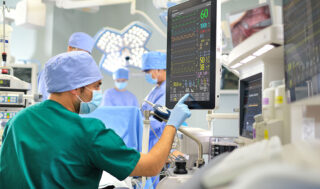
We conduct core research on problems that cut across the data sciences and engineering.
Learn More
We develop analytical and computational tools to manage risk and to support decisions using the growing volume and variety of data available.
Learn More
We use data generated by people and data about people to understand human behavior.
Learn More
We develop the capacity to keep data secure and private throughout its lifetime.
Learn More
We explore the design, analysis, and application of massive-scale computing systems for processing data.
Learn More
We address challenges posed by our data-rich society through clusters of multidisciplinary researchers.

We seek recent Ph.D. graduates with explicit interests in advancing and/or applying data science to other domains.

We support research collaborations between data scientists and domain experts.
